
College of Veterinary Science, Assam Agricultural University, Guwahati-781022, India

| Home | | Check Serotype | | Admin | | View Existing Codes | | About |
Association of Salmonella with serotypes based on Multiplex PCR Genome Typing (MPGT)
This multiplex PCR assay was developed for identification of common clinical serovars of Salmonella enterica and consists of four multiplexes, each one with 4 to 6 gene products (63 to 239 bp). Each of the 16 different target genes shows variability in their presence in different common clinical serotypes and hence the assay can be used for a rapid identification of Salmonella serovars without the need of antisera. Each of the four multiplexes also contains of a universal Salmonella-specific small (63 bp) amplicon, used as an internal control. Users may visit the About page to view the primer sequences and the PCR conditions used.
The genes included in each of the multiplexes are arranged in descending order of corresponding product size. Based on presence or absence of a gene in a multiplex, a corresponding score of 1 or 0 is given (STM1608, which is part of all four multiplexes, has been pre-checked as this gene is present in all Salmonella isolates). In the majority of cases, the resulting MPGT code presents a distinct identification code for a given Salmonella serovar. A few serovars may have more than one MPGT codes due to intra-serovar variations in target gene presence status.
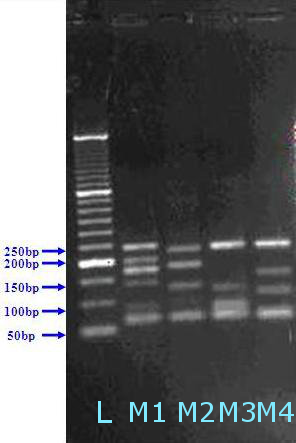 Fig: PCR amplification of all 16 genes included in 4 multiplexes (M1 to M4) against 50bp DNA ladder (L). | In order to use the server to quickly identify the serovar based on the MPGT code, a user only has to indicate the successfully amplified products of his isolate in question by clicking on the corresponding text boxes and submit the query. All submissions made by a registered user are stored in the server for future references. Presently the database consists of reference data from 32 clinically important Salmonella serovars, their MPGT codes and the corresponding antigenic properties. It also contains predicted MPGT codes of 91 Salmonella serovars based on in-silico analysis of 440 Salmonella genome sequences available as NCBI Bioprojects. The database will expand over time with continuous use and incorporation of data from more and more serovars. |